
Start Guide for AccuCal™
Is my study suitable for using AccuCal™ and RealCount™ software?
- Is it a Gene expression study (i.e. measuring mRNA, not miRNA etc)?
- Does your qPCR master mix contain a ‘hot-start’ polymerase?
- Was your cDNA synthesis generated using oligo dTs?
- Are your primer sets generate regular sigmoidal curves?
- Would your qPCR product result in a single dsDNA species (i.e. one band on a gel)?
If the answer is yes to all the above, then your study is suitable for AccuCal™ and RealCount™!
Recommendations for AccuCal™ use:
- We recommend that each sample be run in triplicates. This allows you to detect outliers (if they exist) and improve the accuracy of your results.
- If the primer set efficiency is known for a given master mix, use primer sets with and efficiency of 90%< x<105%. If primer set efficiency is not known, use primer sets that give rise to regular sigmoidal curves. RealCount™ will calculate the PCR efficiency for a given amplicon.
- Optimize qPCR master mix and qPCR thermocycling protocols to get the best sigmoidal amplification curves.
- Amplicons 100-300bp preferred, although AccuCal™/RealCount™ has been robustly tested on amplicons ranging from 70-1000bp. Choose the amplicon length according to your master mix recommendations.
- It is recommended to set up your plate with many different samples but only a few different assays, with as similar product length as possible. This is to guarantee that all 5 parts of the sigmoidal curve for all amplicons are present on the plot.
- For best results it is recommended you use master mixes with a high-resolution melt (HRM) dye, e.g. EvaGreen, BRYT green, SYTO82. These types of dyes do not inhibit the PCR reaction and you will have a sufficient amount of dye to generate a good signal and have reliable PCR performance1.
If your qPCR instrument has user-defined gain and your plate contains assays with disparate length, ensure that the final plateau phase of the sigmoidal curve (maximum fluorescence) of the longest amplicon is detectable on the fluorescence vs cycle number plot.
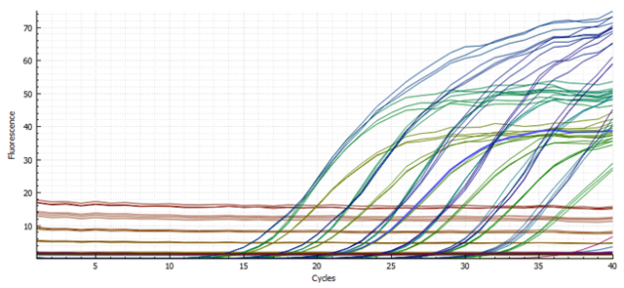
Example of sigmoidal amplification curves and AccuCal standards
What you need before running experiment with AccuCal™
- Install RealCount™ on a windows PC or Mac.
- Raw data (not baseline corrected by the instrument) .rex files can be directly uploaded to RealCount™ or any other format raw data can be exported using provided user friendly Excel template that will contain sample names, fluorescence and cycle-number information.
Advantages of AccuCal™ use
- Every qPCR well on the plate is a complete qPCR assay with individual efficiency and provides quantitative result.
- No dilution series/standard curves of samples with known template concentration.
- Use only 12 to 18 well of prediluted standards per plate.
- Can be used on your existing instrument with master mix of your choice (HRM or SYBR® Green dyes).
- User friendly software that provides you with instant quantitative result.
Limitations of AccuCal™ use
- Must use Hot Start PCR to minimize premature amplification and also delayed onset PCR.
- AccuCal ™ does not currently work with probe-based detection chemistries, such as Taqman probes or hairpin probes.
- AccuCal ™Cannot be applied on miRNA expression research.
- For best results it is recommended using HRM dyes. Dye concentration/reaction can cause inhibition/restriction of SYBR Green dyes1.
Get Started via the Video Demonstrations below!
References:
- Eischeid AC. (2011). SYTO dyes and EvaGreen outperform SYBR Green in real-time PCR. BMC Res Notes. doi: 10.1186/1756-0500-4-263.
